Tree Thinking (3)
In this section, we will explore and discuss the meaning and interpretation of phylogenetic trees estimated from molecular sequence data.
Interpretation of Phylogenetic Trees
In this exercise you will be presented with sets of relatedness relationships between groups of sequences in three different forms: (i) sets of statements; (ii) phylogenetic trees; and (iii) NEWICK format. From each set of relationships that you are presented with, you will then be asked to represent them in a different form.
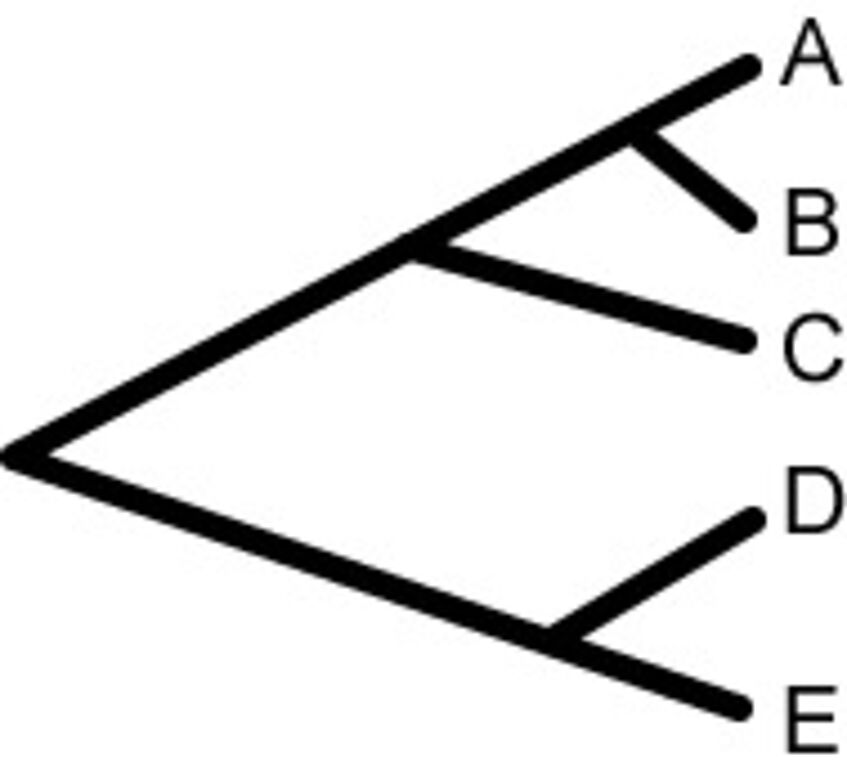
- Provide, for the tree shown, the NEWICK format.
- Based on the three statements given below, provide the corresponding NEWICK format representation of these relationships.
- Sequences A and B are more closely related to each other than either is to any other sequence in the tree.
- Sequence C is more closely related to sequences AB than it is to sequence D.
- Sequence D is equally distantly related to sequences A, B and C.
- Based on the information provided below in NEWICK format, draw the corresponding phylogenetic tree.
(((A,B),((C,D),E)),((F,G),(H,J)))
Unrooted and Rooted Trees
Unrooted trees can be considered as simply representing a set of rooted trees. The process of rooting an unrooted tree is that of placing an "ancestral' node onto the unrooted tree. This node is that of the earliest divergence present in the tree. It provides a temporal order to the divergences represented in the tree - all other divergences in the tree occurred after that of the ancestral node. The number of trees grows exponentially with the number of taxa. To get an idea how the numbers explode look here.
Which of the following trees is rooted and which unrooted?
- ((((A,B),C),D),E,(F,G))
- (((A,B),(C,D)),(E,F))
- (A,(((B,D),C),(F,(E,G))))
Orthologs and paralogs
The aim of this exercise is to give you practise specifying the root of a gene tree by identifying the root that minimises the complexity of the evolutionary scenario required to explain the gene tree. In the exercise you are asked to use a species tree and a gene tree to estimate the root of the gene tree.
For the following gene-tree/species-tree pair containing sequences from five dif ferent vertebrates, someone who you trust assures you that the root of the gene tree is located on one of the internal branches of the tree, when the root is assumed to be that which yields the evolutionary scenario that includes the smallest number of gene duplication events.
(zebrafish,(frog,(mouse,human)));
((mouse,human2),(frog2,human1),(zebrafish2,(frog1,zebrafish1)));
Place the root of the tree at each of the internal branches in turn, and identify that internal branch that, when used as the root, minimises the number of gene duplication events.
