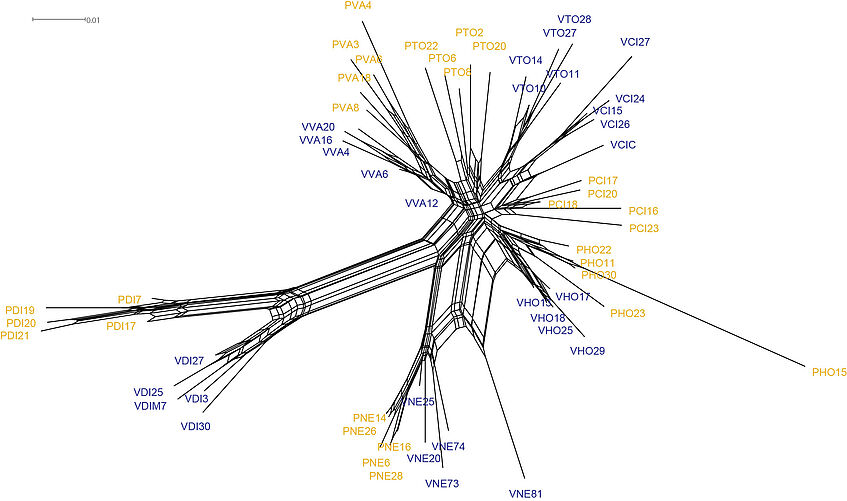
Building an evolutionary network
Our next aim is to use SplitsTree to get a first impression on the evolutionary relationships between individuals. For this we need to transform the .vcf file into a .nex file. We can use for example PGDSpider. Direct conversion results in improper coded SNP format. We need the data to be coded as DNA sequences, i.e., one line per individual and one code per position, by using the IUPAC encoding for ambiguous positions.

Open PGDSpider with:
PGDSpider
First produce from the .vcf a .phylip by selecting “sequence data” and “DNA” to be output in the .phylip. Then produce a .nex from the .phylip by selecting data type “DNA”. Select the input file and give a name for the output file in the correct folder (~/RADlecture/). Create SPID files. Click convert.
To construct the network, open SplitsTree with
SplitsTree
File -> Open and select the .nex file.
- Do the ecotypes form each a unit/cluster? How many origins can we infer of each ecotype based on the network?
- What do the reticulations represent here? Should we expect relationships at the population level rather fit bifurcating trees or networks?